Klotho, one of the three Fates in Greek mythology, spins the thread of human life. Similarly, the Klotho gene affects human longevity and resilience to age-related cognitive decline. In our paper we describe a major perturbation in RNA (including messenger RNA, long non-coding RNA, microRNA and tRNA fragments (tRF)) following Klotho knockout (KO) in mouse brains. Interestingly, many of the altered transcripts and gene programs show changes similar to those occurring in Alzheimer’s Disease and brain aging, suggesting that Klotho KO triggers a cascade of molecular events akin to those in neurodegeneration and age-related cognitive decline. In particular, Klotho KO elevates the expression of stress-induced tRNA halves that interact with the spliceosome, possibly affecting gene expression. Finally, to evaluate the cell-type specificity of Klotho KO-perturbed short non-coding RNAs, we collected live human brain samples extracted during neurosurgery, and sorted the nuclei into those of neurons and microglia, followed by short RNA sequencing. This unique resource revealed that microRNAs are highly cell-type specific, with microglial microRNA signaling reduced and neuronal signaling elevated following Klotho KO, a result that is consistent with mRNA-based observations. In contrast, tRFs emerged as more prominently apparent in neurons. All our data, including the transcriptomic signature of murine brains following Klotho KO and neuron- and microglia-specific short non-coding RNA profiles, are publicly available.

Figure 1
We extracted and sequenced the total RNA from murine brains of wildtype (WT) and Klotho knockout (KL-/-) genotypes (n=5 in each group). This allowed us to identify changes in numerous RNA fragments, including mRNA, long non-coding RNA, microRNA and tRNA fragments. In particular, we sought correlations of the observed transcriptomic changes with reported aging and AD alterations and pursued the cell type-specificity of the observed differences.
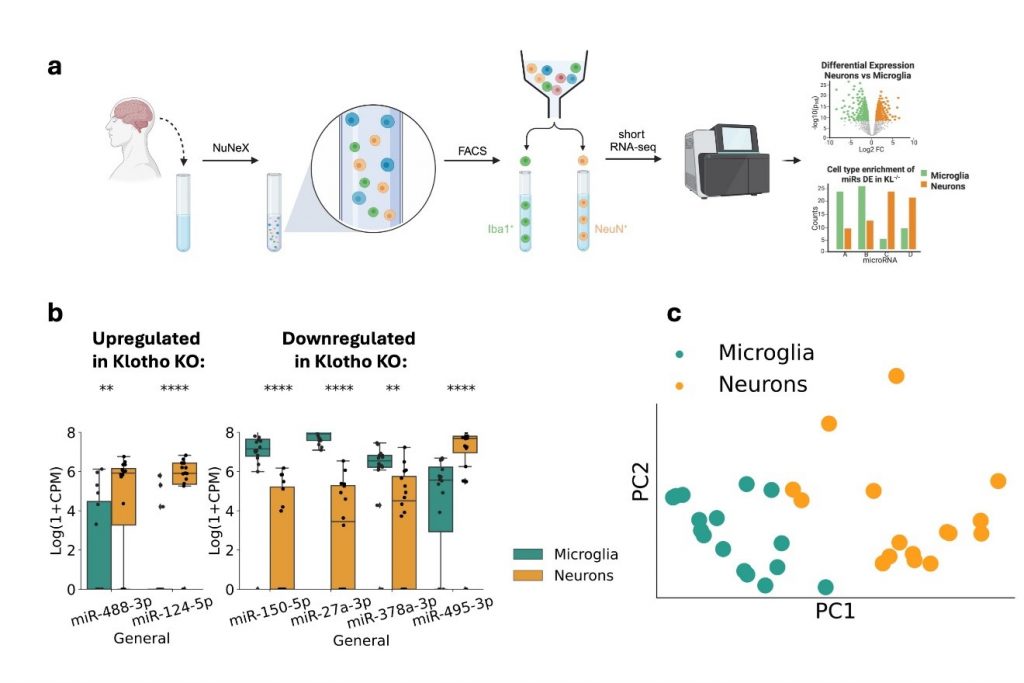
Figure 2
a) Live brain samples extracted during neurosurgeries were broken down to single nuclei surrounded by a thin layer of cytoplasm, using our in-house NuNeX protocol. The nuclei were stained with microglia- (Iba1) and neuron- (NeuN) specific fluorescent markers, followed by FACS sorting and short RNA-sequencing.
b) 6 miRS DE in Klotho KO, are also enriched in one of the cell types. Left: miRs upregulated in Klotho KO, right: miRs downregulated in Klotho KO; **: padj<=0.01; ****: padj<=0.0001.
c) PCA of the generated cell type-specific small transcripts (7 miRs DE in Klotho KO, colored by cell type).


